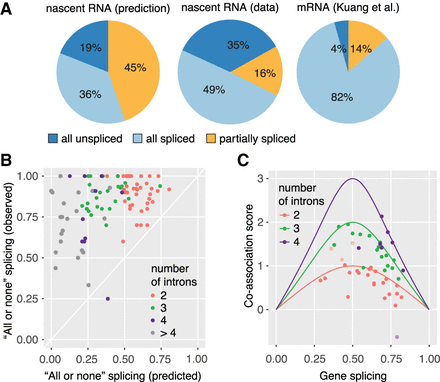
High degree of splicing co-association in multi-intron transcripts. (A) Predicted and observed “all unspliced,” “all spliced,” and “partially spliced” fractions in the transcriptome. Left panel: Prediction of splicing categories from nRNA-seq, assuming each intron is spliced independently of neighbors. Predicted values were calculated for the first two introns of genes with two and more introns for nRNA. Middle panel: Proportion of the three splicing categories from nRNA LRS as in Figure 3B. Right panel: Proportion of the three splicing categories from total mRNA LRS (data from Kuang et al. 2017). (B) Predominant “all or none” splicing in multi-intron genes. The predicted fraction of “all or none” splicing (fraction of all spliced or unspliced transcripts calculated from nRNA-seq, assuming splicing independence) is plotted against the observed fraction of “all or none” splicing for 100 genes with 10 or more LRS transcripts. (C) Co-association of splicing is close to the maximal possible co-association value. A co-association score was calculated as the log2-fold change of the observed (LRS) to predicted (RNA-seq) fraction of “all or none” splicing. The maximal possible co-association score for 2-, 3-, and 4-intron genes was calculated [log2(1/gene splicing)] and plotted versus the mean gene splicing value (solid line). Co-association scores for individual genes fall below or on top of this line, suggesting maximal co-association for most genes (median 86%, calculated as the ratio of the observed co-association score over the maximal co-association score at a particular gene splicing). Outliers are indicated as lighter points and can be explained by inconsistencies in gene annotation.











