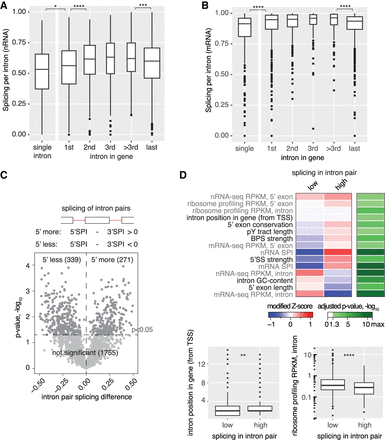
Pre-mRNA features that correlate with the extent of cotranscriptional splicing. (A) Cotranscriptional splicing levels differ based on gene position. The box plot shows the distribution of nRNA SPIs for the group of single intron genes and first, internal (second, third, and other), or last introns in multi-intron genes. The box width corresponds to the respective group size. (B) mRNA splicing levels differ between introns in different gene positions. Analogous data representation as in A. (C) One quarter of introns are significantly less or more spliced than the next downstream (3′) intron in nRNA, as depicted in a volcano plot (three biological replicates). (D) 15/42 analyzed gene architecture features correlate significantly with differentially spliced intron pairs (sequence-based in black font and RNA-seq-derived in gray font). The smaller intron position for “low” spliced introns in a pair (first intron – 1, second intron – 2, etc.) is consistent with enrichment of first introns in the “5′ less spliced” group. The median modified Z-score is shown for each feature with significant difference between the “low” and “high” groups, and the respective negative log10 of the Bonferroni-corrected P-value is given. For two features, no change in the median modified Z-score is visible. The respective feature distribution is presented as a box plot below. Asterisks indicate significance of direct neighbors according to the Wilcoxon rank-sum test: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001 after Bonferroni-correction, in A, B, and D.











