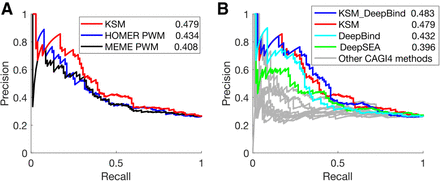
Figure 6.
KSMs predicts allele-specific differences in regulatory activity better than PWMs and deep learning–derived features. (A) PRC performance of KSM and PWM motif representations in predicting differential regulatory activities of eQTL alleles. The numeric values in the legend are the AUPRC values. (B) Similar to A, KSM, DeepBind, and DeepSEA derived features and other CAGI 4 open challenge methods.











