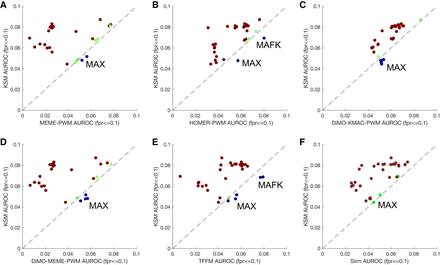
KSMs outperform PWMs and complex motif models in predicting in vitro TF binding. Scatter plots compare the mean partial AUROC performance of KSM versus MEME PWM (A), HOMER PWM (B), DiMO-optimized KMAC PWM (C), DiMO-optimized MEME PWM (D), TFFM (E), and Slim (F) motif models for predicting HT-SELEX in vitro TF binding. Each point represents a ChIP-seq experiment of which the TF has been profiled using HT-SELEX. (Brown) KSM performs better than other motif representations; (blue) KSM performs worse; (green) both representations perform similarly.











