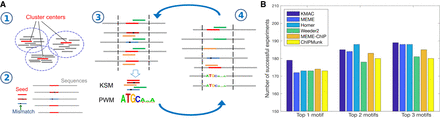
KMAC motif discovery outperforms other methods when detecting motifs in ChIP-seq data. (A) KMAC motif discovery schematic. Step 1: Overrepresented k-mers with length k are clustered using density-based clustering. Bars represent the k-mers, whereas red bars represent the cluster center exemplars. Step 2: A cluster center is used as a seed k-mer. The seed k-mer and k-mers with a one-base mismatch are used to match and align the sequences. Step 3: A pair of KSM and PWM motifs are extracted from the aligned sequences. Step 4: The KSM and PWM motifs are used to match and align the sequences. Steps 3 and 4 are repeated until the significance of the motifs stops to improve. (B) The motif discovery performance of KMAC is compared to the motif discovery performance of various motif finders on 209 ENCODE ChIP-seq experiments.











