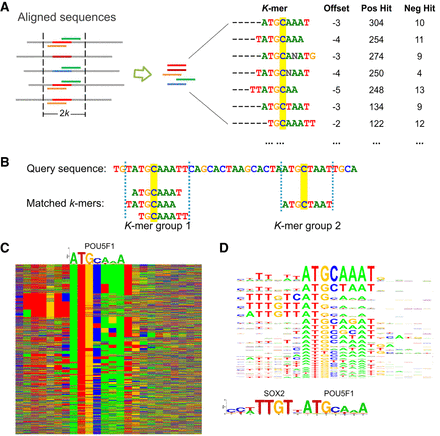
The KSM motif representation. (A) A KSM consists of a set of similar and consistently aligned component k-mers. The k-mers are extracted from a set of sequences aligned at the binding sites. Each k-mer has an offset that represents its relative position in the sequence alignment and is associated with the IDs of the positive/negative training sequences that contain the k-mer (IDs are not shown, total counts are shown). The base C, highlighted in yellow, represents the expected binding position. (B) An example of matching KSM motifs in a query sequence. (C) Color chart representation of 2183 sequences bound by POU5F1 that match the POU5F1 KSM motif. Each row represents a 23-bp sequence. Rows are sorted by the KSM motif matches. Green, blue, yellow, and red indicate A, C, G, and T, respectively. A POU5F1 PWM motif is shown above the sequences. (D) The KSM motif sequence logo of POU5F1 (corresponding to the aligned sequences in C) and the PWM logos of SOX2 and POU5F1.











