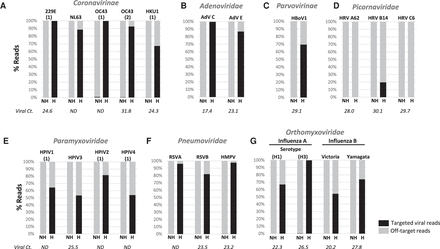
Distribution of sequence reads for clinical samples enriched with virus-specific probes. The frequency of reads identified by Kraken for each sample with and without enrichment (H:hybridized; NH: nonhybridized) is shown in bar graphs for each viral family/subfamily tested: (A) Coronavirinae; (B) Adenoviridae; (C) Parvovirinae; (D) Picornaviridae; (E) Paramyxoviridae; (F) Pneumoviridae; (G) Orthomyxoviridae. Viral Ct is shown below the bar graphs. (ND) Ct not available. Abbreviations of virus names are listed in Supplemental Table S1.











