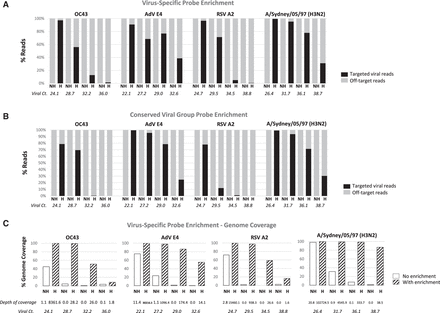
Sensitivity of enrichment in hybridization. Samples were prepared as 10-fold serial dilutions of reference viral nucleic acids spiked into a constant amount of human RNA prior to library preparation. The frequency of reads identified by Kraken is shown in bar graphs for each sample with and without enrichment (H: hybridized; NH: nonhybridized) for (A) virus-specific probe enrichment and (B) conserved viral group probe enrichment. From the same sequencing run, the linear genome coverage is shown (C) for samples with enrichment (diagonal stripes) or without enrichment (white) with virus-specific probes. Viral Ct and (average) depth of coverage are shown below the bar graphs. Abbreviations of virus names are listed in Supplemental Table S1.











