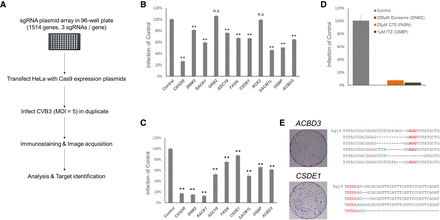
Candidate identification. (A) Schematic presentation of the arrayed CRISPR screen. (B) Quantification of CVB3 infection in HeLa cells transfected with sgRNA-expressing plasmids targeting 10 genes from primary screens. Mean ± SEM for quadruplicate experiments. (**) P < 0.01, (n.s.) nonsignificant. (C) Similar quantification of virus infection in B using newly designed sgRNA. (D) The percentage of CVB3-infected cells (normalized to DMSO control) treated with three known inhibitors of the candidates. Mean ± SD for triplicate experiments. (E) HeLa cells were transfected with ACBD3 and CSDE1 targeting sgRNAs and infected with CVB3. Surviving cells were stained and are shown. Surviving colonies were expanded and the target region was sequenced to confirm the mutations. Red characters indicate the PAM and blue characters indicate insertions. hg19 is the wild-type sequence of each gene. As our target sites have the same sequence in both references (i.e., hg19 and GRCh38), using GRCh38 would not significantly affect our conclusions.











