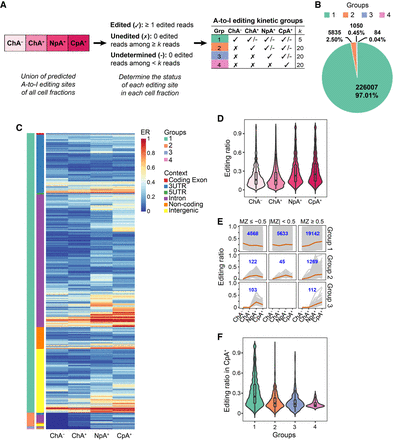
Kinetic groups of RNA editing sites identified via subcellular RNA-seq. (A) Categorization of RNA editing sites into four kinetic groups using cell fractionation RNA-seq. Editing sites in each group are labeled as edited (✓), unedited (✗), or undetermined (-) in each subcellular fraction. The read coverage cutoffs of the editing sites (k) are illustrated in the table (for details, see Methods). (B) Number and percentage of A-to-I RNA editing sites in each editing kinetics group. (C) The editing ratios (ER) of each editing site in the four groups. The genomic location of an editing site is shown in color codes. All editing sites included in this analysis were required to have at least five total reads in each fraction (same for panels D–F). A total of 29,345, 1437, 220, and 84 sites were included for groups 1, 2, 3, and 4, respectively, with group 1 editing sites accounting for 94% of all. (D) Editing ratio distribution of the editing sites in C (union of all four kinetic groups) in each subcellular fraction. Note that for each fraction, editing ratios of the union of all groups are shown. (E) Editing ratios in different subcellular fractions of editing sites in C with different ranges of MZ scores. Each gray line represents one editing site. The average editing ratios are highlighted in orange. The number of editing sites in each panel is shown in blue. It should be noted that MZ scores cannot be calculated for sites with constant editing ratios across two consecutive fractions; thus, they were excluded from this analysis. (F) Editing ratios of each kinetics group observed in mature mRNAs (CpA+).











