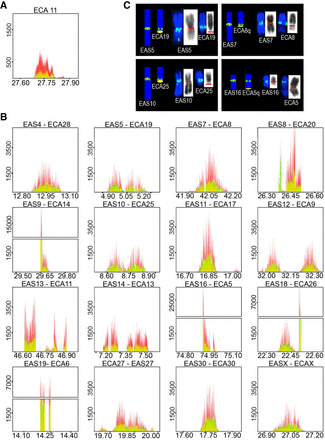
Identification of satellite-free centromeres in Equus asinus. ChIP-seq reads from primary fibroblasts of HorseS (A) and DonkeyA (B) were mapped on the EquCab2.0 horse reference genome. Immunoprecipitation was performed with an antibody against human CENPA (red) or with a CREST serum (green). Peak overlapping appears in yellow. The y-axis reports the normalized read counts, whereas the x-axis reports the genomic coordinates (Mb). The E. caballus satellite-free centromere from Chromosome 11 (A) and the 16 satellite-free E. asinus centromeres (B) are shown; for each E. asinus (EAS) chromosome, the number of the orthologous E. caballus chromosome (ECA) is reported. (C) FISH with BAC probes covering the genomic regions identified by ChIP-seq. Four examples (EAS) along with their orthologous horse chromosomes (ECA) are shown; the remaining chromosomes are reported in Supplemental Figure S1. On the left of each panel, a sketch of the orthology between E. caballus and E. asinus chromosomes (Yang et al. 2004; Musilova et al. 2013) is shown, with BAC signals represented as green dots, and the position of the cytogenetically determined primary constriction represented as a yellow oval. On the right of each panel, metaphase chromosomes are shown with FISH signals in green, and the primary constriction is marked by a red line on the reverse DAPI images (gray).











