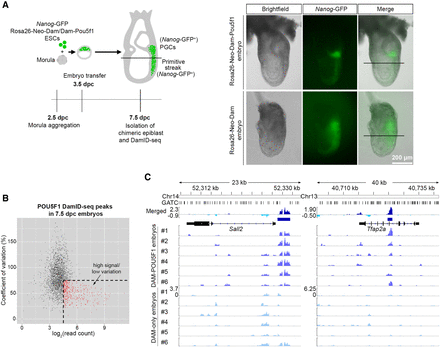
POU5F1 DamID-seq with 7.5-dpc epiblasts. (A) DamID-seq samples were prepared from each Dam/Dam-POU5F1–expressing 7.5-dpc chimeric embryo generated via morula aggregation, excluding the PGC-containing region. Nanog-GFP confirms contribution of Dam/Dam-POU5F1 ESCs, while the reporter expression is limited to posterior. (B) Read counts and coefficient of variation of the epiblast POU5F1 DamID-seq peaks. Peaks with high read counts (log2 > 4.5) and low standard deviation (<75%) indicated in red were used for further analyses. (C) The merged epiblast POU5F1 DamID-seq tracks (top) generated from six Rosa26-Neo-Dam-Pou5f1 and six Rosa26-Neo-Dam embryos. Blue bars indicate selected confident peaks from B.











