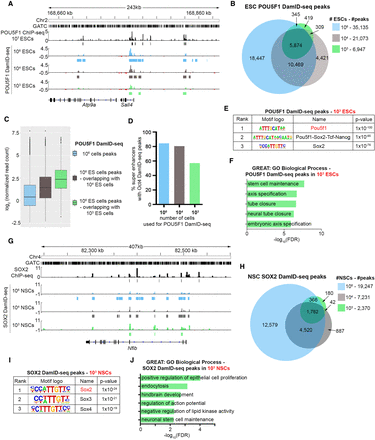
104, 103 ESC POU5F1 DamID-seq and 104, 103 NSC SOX2 DamID-seq. (A) POU5F1 DamID-seq tracks from 106/104/103 ESCs and POU5F1 ChIP-seq track from 107 ESCs (Buecker et al. 2014). (B) Overlaps of 106/104/103 ESC POU5F1 DamID-seq peaks. (C) Read counts of peaks in 106 ESC DamID-seq (blue) and those identified by 104 (gray) and 103 (green) ESC DamID-seq. (D) Percentage of ESC super enhancers (Whyte et al. 2013) containing POU5F1 DamID-seq peaks using different number of cells. (E) Motif enrichment in the 103 ESC POU5F1 DamID-seq peaks. (F) GO enrichment analysis of 103 ESC POU5F1 DamID-seq peaks using GREAT (McLean et al. 2010) (G) SOX2 DamID-seq tracks from 106/104/103 NSCs and SOX2 ChIP-seq tracks generated from 5 × 106 NSCs (Mateo et al. 2015). (H) Overlaps of 106/104/103 NSC SOX2 DamID-seq peaks. (I) Motif enrichment in the 103 NSC SOX2 DamID-seq peaks. (J) GO enrichment analysis of SOX2 DamID-seq peaks from 103 NSCs.











