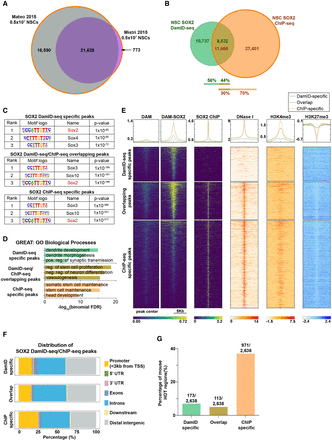
106 NSC SOX2 DamID-seq in comparison with ChIP-seq. (A,B) Union of two published NSC SOX2 ChIP-seq data sets (A) (Mateo et al. 2015; Mistri et al. 2015) and its overlap with 106 NSC SOX2 DamID-seq peaks (B). (C) Motif enrichment analysis (Heinz et al. 2010) of the SOX2-bound peaks identified only by DamID-seq, ChIP-seq, or both technologies. (D) GO enrichment analysis of SOX2 peaks identified only by DamID-seq, ChIP-seq, or both using GREAT (McLean et al. 2010). (E) For each SOX2 DamID/ChIP-seq overlapping and specific peaks, the signal intensities from SOX2 DamID-seq (Dam; Dam-SOX2 separately), SOX2 ChIP-seq, DNase-seq, and H3K4me3 and H3K27me3 ChIP-seq are represented. (F) Distribution of SOX2 DamID/ChIP-seq overlapping and specific peaks in the genome. (G) DamID/ChIP-seq overlapping and specific peaks including mouse ChIP-seq HOT regions (Wreczycka et al. 2017).











