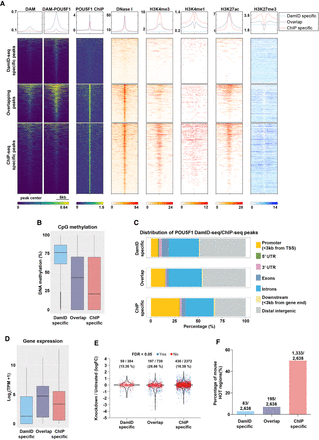
Characterization of POU5F1 DamID-seq and ChIP-seq peaks. (A) For each POU5F1 DamID/ChIP-seq overlapping and specific peaks, the signal intensities from POU5F1 DamID-seq (Dam; Dam-POU5F1 separately), POU5F1 ChIP-seq, DNase-seq, and H3K4me3, H3K4me1, H3K27Ac, and H3K27me3 ChIP-seq are represented. (B) CpG methylation levels of POU5F1 DamID/ChIP-seq overlapping and specific peaks. The ChIP-seq–specific peaks were extended to the same average size of DamID-seq peaks as to avoid biases due to different size of the peaks. (C) Distribution of POU5F1 DamID/ChIP-seq overlapping and specific peaks in the genome. (D) Expression level of genes with DamID-seq–specific (384 genes), overlapping (739 genes), and ChIP-seq–specific peaks (2372 genes) in their promoter regions (±3 kb from TSS). (E) Expression changes of genes in D, 24 h after knockdown of Pou5f1 in ESCs (King and Klose 2017). (F) DamID/ChIP-seq overlapping and specific peaks including mouse ChIP-seq HOT regions (Wreczycka et al. 2017).











