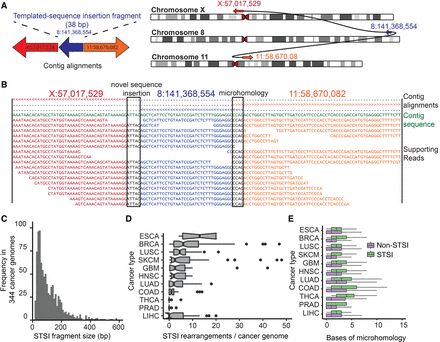
SvABA identifies rearrangements with short templated-sequence insertions (STSI) derived from distant genomic loci. (A) Somatic rearrangement between Chr X and Chr 11 in HCC1143 containing a 38-bp fragment of Chr 8. STSI rearrangements are identified by assembly contigs that have multiple non-overlapping alignments to the reference. The direction of the arrows represents the strand that the contig fragment was aligned to (right-facing is forward strand). (B) Partial view of the contig from A showing the multiple alignments of the contig to the reference and the read-to-contig alignments. The top three lines indicate which bp of the contig each of the three BWA-MEM alignments covers (> is forward strand alignment; < is reverse strand alignment). The first two alignments indicate an insertion of 5 bp of novel sequence at the first junction (left), and the second two indicate 4 bp of microhomology at the second junction (right). The middle alignment supports the STSI fragment. These plots are automatically generated by SvABA for each variant (in the *.alignments.txt.gz file). (C) STSI fragment lengths from somatic rearrangements across 344 cancer genomes (mean 86 bp). (D) Prevalence of STSI rearrangements (x-axis) across 11 tumor types (y-axis). (ESAD) esophageal cancer; (BRCA) breast cancer; (LUSC) lung squamous cell carcinoma; (SKCM) melanoma; (GBM) glioblastoma; (HNSC) head and neck squamous cell carcinoma; (LUAD) lung adenocarcinoma; (COAD) colorectal adenocarcinoma; (THCA) thyroid carcinoma; (PRAD) prostate adenocarcinoma; (LIHC) hepatocellular carcinoma. (E) Bases of breakpoint microhomology (x-axis) for different cancer types (y-axis) for somatic STSI rearrangements (green) and somatic non-STSI rearrangements (purple). The STSI rearrangements have a significantly higher degree of breakpoint microhomology than their non-STSI counterparts across all tumor types.











