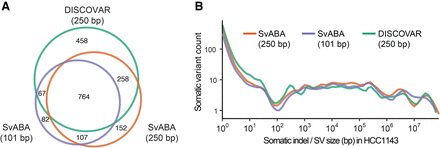
Somatic variant detection in the HCC1143 breast cancer cell line using different sequencing and informatics approaches. (A) Comparison of combined somatic SV and indel detection in HCC1143 using: local assembly using SvABA with 101-base paired-end reads (purple), SvABA with 250-base paired-end PCR-free reads (orange), or global assembly using DISCOVAR de novo on 250-base paired-end PCR-free reads and SVlib to extract variants (green). (B) Somatic variant counts (y-axis) for DISCOVAR de novo (250-base PCR-free reads; green) and SvABA using 101-base (purple) or 250-base PCR-free reads (orange), as a function of variant size (x-axis). All methods have similar sensitivities across different sizes, except DISCOVAR de novo was more sensitive to short indels in simple repeats.











