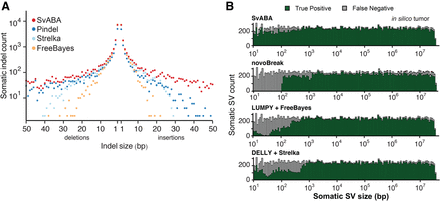
Benchmarking somatic variants with an in silico tumor. (A) True positive counts for indel calling (y-axis) as a function of variant size (x-axis) for SvABA (red), Pindel (blue), FreeBayes (orange), and Strelka (light blue). All callers achieved similar sensitivities for small somatic indels, while SvABA maintained high sensitivity for larger (>10 bp) indels. (B) Stacked bar chart of the number of SVs detected across all SV types (y-axis) as a function of variant size (x-axis). SvABA maintained sensitivity across variants of all sizes. novoBreak had the second highest sensitivity for medium and large variants after SvABA. Combining calls from a dedicated indel and SV caller (LUMPY and FreeBayes or DELLY and Strelka) improved overall sensitivity, but still left a gap for medium-sized SVs.











