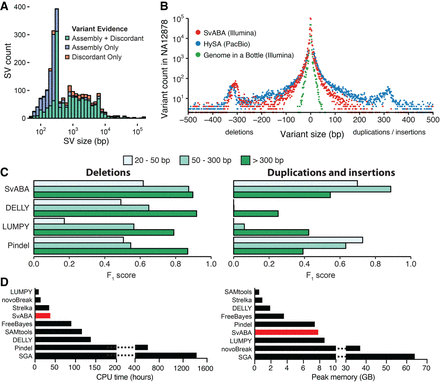
Detection of SVs and indels in the NA12878 human genome. (A) The number of SV events and the types of supporting evidence used by SvABA for detecting SV events of different lengths (indel variants not shown). SVs are detected through realignment of assembly contigs (purple), discordant read clusters (orange), or a combination of both (green). SVs with shorter lengths than the average size of the sequencing fragments are identified almost exclusively through assembly and realignment. (B) The length distributions of indels and small SVs in NA12878 determined from different sequencing and analytical technologies: 151-base paired-end Illumina sequencing by SvABA (red), HySA calls from PacBio sequencing data (blue), and the indel call set of the Genome in a Bottle consortium (green). (C) Comparison of detection accuracy of SvABA, LUMPY, DELLY, and Pindel for deletions (left) and for insertions/duplications (right) across three different length regimes in NA12878. The F1 score is a combined measure of precision and recall and was calculated using the PacBio assemblies and Genome in a Bottle (GIAB) as a truth set. (D) Total CPU and peak memory usage for several indel and SV detection tools applied to a single 33× human genome. SGA CPU and memory usage were estimated using published data (Simpson and Durbin 2012).











