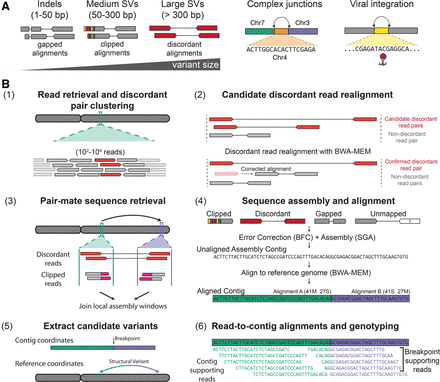
Overview of the SvABA structural variation detection tool. (A, left) SvABA uses String Graph Assembler (SGA) to assemble aberrantly aligned sequence reads that may reflect an indel or SV. Such reads include gapped alignments (for indels), clipped alignments (for medium and large SVs), and discordant read pairs (for large SVs). In addition to detecting indels and SVs, SvABA can identify complex rearrangement junctions (middle) and sites of viral integration (right). (B) The workflow for the SvABA pipeline: (1) reads within a small window are extracted from one or multiple BAM files and discordant reads are clustered; (2) discordant reads are realigned to the reference to remove pairs that have a candidate nondiscordant alignment; (3) the discordant read clusters are used to identify additional regions where reads should be extracted; (4) the sequences are error-corrected with BFC and assembled with SGA into contigs, and contigs are immediately aligned to the reference with BWA-MEM; (5) contigs with multipart alignments or gapped alignments are parsed to extract candidate variants; and (6) sequence reads are aligned to the contig and to the reference to establish read support for the reference and alternative haplotypes.











