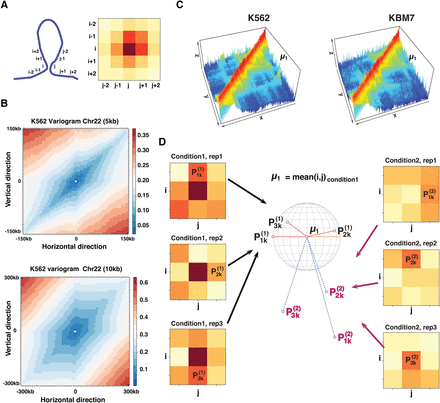
Existence of the spatial dependency and the idea behind our model. (A) Illustration of the spatial dependency along neighboring loci in the Hi-C contact map. (B) Semivariogram showing the directional variability between interaction bins separated by a certain horizontal and vertical
distance in the Hi-C contact map. (C) A differential interaction can be considered as a change of the intensity around the 3D coordinate (i, j, fij) of a reference point μ1. (D) Principle of the k-nearest neighbor (KNN) intensity estimation in a 3×3 window around a pairwise interaction (i, j). Given an interaction (i, j) with a mean frequency 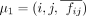 in the first condition (represented by the mountain tip in C), if there is no structural change, we expect the interaction frequencies from the second condition to have a similar density
around the point μ1 in the 3D space. Thus, for each condition and each replicate, we calculate the 3D distance between each bin in the surrounding
window and μ1 and order them according to their distance. We note
in the first condition (represented by the mountain tip in C), if there is no structural change, we expect the interaction frequencies from the second condition to have a similar density
around the point μ1 in the 3D space. Thus, for each condition and each replicate, we calculate the 3D distance between each bin in the surrounding
window and μ1 and order them according to their distance. We note  as the kth-nearest neighbor to the point μ1 from the nth replicate of condition c ∈ {1, 2}. Then, we estimate the density of the KNNs around μ1in the first condition
as the kth-nearest neighbor to the point μ1 from the nth replicate of condition c ∈ {1, 2}. Then, we estimate the density of the KNNs around μ1in the first condition 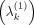 and in the second condition
and in the second condition  . The density of the KNNs around μ1 is expected to be stable between the two conditions. To decide if the change of the KNN density around μ1
. The density of the KNNs around μ1 is expected to be stable between the two conditions. To decide if the change of the KNN density around μ1  is significantly large or small, we use a Fisher distribution. The same principle applies if we use μ2 (the mean in condition 2) as our reference point.
is significantly large or small, we use a Fisher distribution. The same principle applies if we use μ2 (the mean in condition 2) as our reference point.











