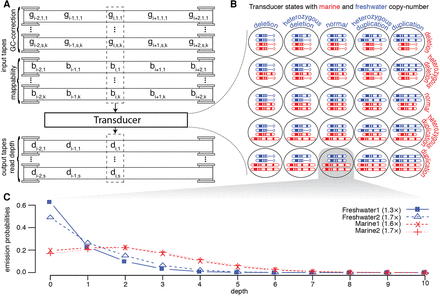
The transducer model. (A) The transducer reads from s · k+k tapes containing the GC-bias and mappability of all overlapping reads, where s is the total number of individuals being analyzed, and k is the length of sequencing reads. gi,n,j is the GC-bias of the jth of k read position that would cover assembly position i for sample n of s. bi,j is the mappability of the jth of k read position that would cover position i. There are multiple output tapes, one for each individual, containing the read depth for that sample. di,n is the depth of reads at assembly position i for sample n of s. (B) The 25 states represent the variation in canonical copy number that can be detected by the transducer. (C) One example of the emission probabilities for a single base with the state representing no change in freshwater copy number, while the marine fish have a duplication. We show the marginal probability distribution for four individuals.











