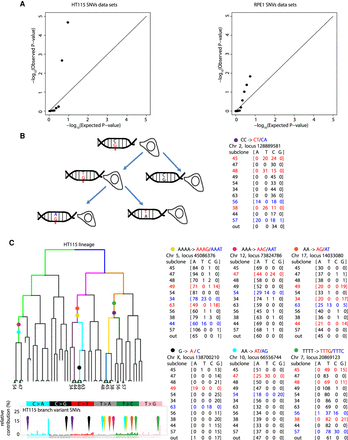
Intra-lineage heterogeneity in mutation rate and multiple mutation events. (A) Measured P-values from observed mutation counts are plotted vs. calculated theoretical (Poisson) P-values for the branch variant set for each sublineage to form quantile-quantile (QQ) plots (P-values for theoretical Poisson-distributed lineage-wise mutation count data versus observed data) for both HT115 (left) and RPE1 (right). The plotted points deviate strongly from the expected distribution (which would follow an x = y relationship) at both ends of the distribution, showing that the sublineages present in each data set cannot be plausibly modeled by a Poisson distribution based on independent mutations. (B) Schematic showing persistent lesion hypothesis for correlated same-site mutation. DNA lesions (marked as G*) that are not repaired during S-phase compel the DNA polymerase to replicate opposite lesion bases with a high probability of mutation. If the lesion has not been repaired before the next S-phase in the daughter cell carrying the lesion, an additional mutation at the identical genomic locus is likely to result. If the second mutation is different from the first, this process can be readily detected from lineage sequencing data. The example scheme represents the CC > CT and CC > CA mutations we detected at the Chromosome 2 locus 128889581 (marked with purple circle). (C) Seven multiple mutation events were found in the HT115 lineage. Read counts for each example are presented and marked with a colored symbol. The lineage segment where each example occurred is shown (with corresponding colors). None of these events overlap the most probable mutation types found in the POLE signature.











