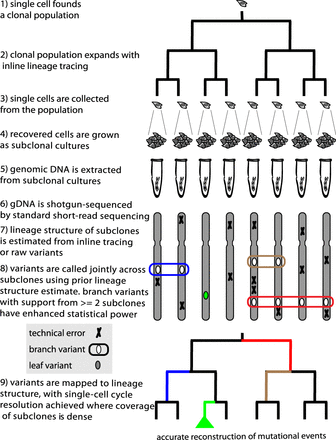
Lineage sequencing concept and implementation. Overview of the lineage sequencing concept. Numbering indicates key conceptual and implementation steps. Single cells are sampled from a clonal population and sequenced (steps 1–6; in this study, subclonal culture was used to produce enough genomic DNA for PCR-free shotgun sequence library construction). Crucially, a prior estimate of the population lineage structure (step 7; either from single-cell tracking by time-lapse imaging or from raw SNV calls) was used to identify novel somatic variants in a joint analysis of the sequence libraries (step 8). This use of the lineage information enables all the sequence libraries to provide statistical support for somatic “branch variants,” enhancing the sensitivity and specificity of somatic variant identification (for example, the schematically indicated “red variant” is supported by presence of an SNV in four sequence data sets from one sublineage and the absence of this SNV in four additional sequence data sets from the other sublineage). Where the coverage by sampled sublineages (via subclones in this study) is high, the mutations that appeared during the lineage experiment can be mapped with single-cell resolution onto the lineage (step 9; e.g., blue, red, tan segments in the dendrogram at bottom). We term mutations occurring in the last round of cell division events “leaf variants,” which by definition can be supported only by a single sequence data set (e.g., green segment in the dendrogram at bottom). Leaf variants can also be analyzed but do not benefit from the enhanced statistical power that supports branch variants and thus cannot be reliably assigned to specific cellular events.











