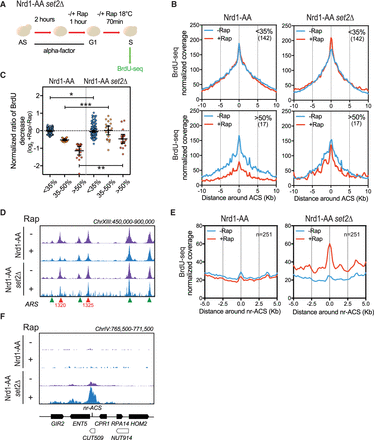
Absence of Set2 H3K36 methyltransferase partially rescues replication defects due to noncoding transcription readthrough. (A) Experimental scheme as described in Figure 1C. (B) Metagene analysis of the BrdU-seq for the 142 nonaffected ARSs (<35%) and the 17 most affected ARSs (>50%) for the Nrd1-AA and Nrd1-AA set2Δ strains. Plots for the Nrd1-AA strain were already presented in Figure 1F, with the exception of the normalized coverage, for which calculation is detailed in Methods. Profiles represent the mean coverage smoothed by a 200-bp moving window. ARSs were oriented according to their ACS T-rich sequence. (C) Scatter dot-plot presenting the normalized BrdU ratio for the different classes of ARS affected in BrdU incorporation in an Nrd1-AA strain and for the same classes of ARSs in the Nrd1-AA set2Δ strain. (D) Snapshot depicting the BrdU-seq reads for a part of Chromosome XIII. ARSs that are rescued in BrdU incorporation in the Nrd1-AA set2Δ +Rap condition are depicted in red, while nonaffected ARSs are in green. (E) Metagene analysis of BrdU incorporation 5 kb around nonreplicating ACSs (nr-ACS) in the indicated strains grown in −/+Rap. The representation is smoothed over a 200-bp moving window. (F) Snapshot illustrating the activation of a dormant nr-ACS in the Nrd1-AA set2Δ +Rap condition. (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001.











