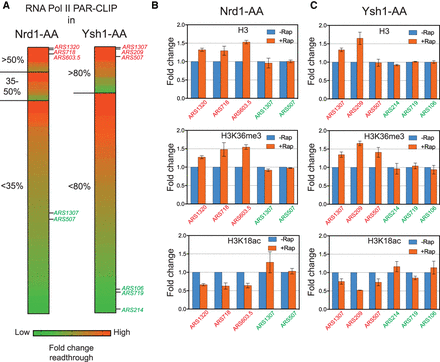
Noncoding and mRNA transcription readthrough at replication origins leads to chromatin changes. (A) Heat map representing the fold change of transcriptional readthrough at replication origins in Nrd1-AA and Ysh1-AA mutants. The 178 ARSs were classified according to their BrdU incorporation defects and ranked by their total readthrough increase over the ACS to +100 bp region. The three ARSs indicated in red for each mutant present a high increase in total readthrough. The ARSs depicted in green show mid or low total readthrough and have been used as controls for the following experiments. (B,C) Chromatin immunoprecipitation (ChIP) of H3, H3K36me3, or H3K18ac at ARSs with high (red) and low (green) readthrough transcription. Asynchronous cells were treated 1 h or 30 min with rapamycin to induce Nrd1 and Ysh1 depletion from the nucleus, respectively. ChIP was performed as described in Methods. Immunoprecipitated ARS loci were normalized to immunoprecipitated SPT15 ORF after qPCR amplification. Fold enrichment was artificially set to 1 for the −Rap condition (n = 3). Error bars represent the standard error of the mean (SEM).











