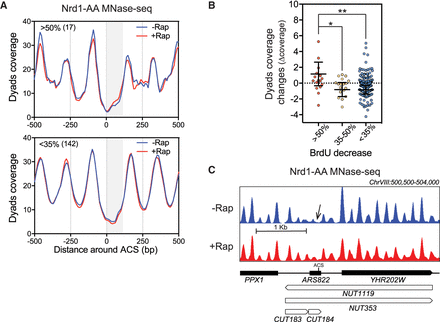
Noncoding transcription readthrough into replication origins alters nucleosome occupancy. (A) Metagene analysis of MNase-seq from Nrd1-AA cells treated or not with rapamycin for 1 h at the three classes of ARSs defined in Figure 1. Only paired-end reads from 120 to 200 bp length were considered to define nucleosome dyad coverage. The gray box represents the window in which transcriptional readthrough was analyzed. (B) Scatter dot-plot representing the difference of dyads coverage (Δcoverage = coverage [+Rap] − coverage [−Rap]) between the ACS to +100 of oriented ARSs when comparing +Rap and −Rap conditions. (C) Snapshot of the MNase-seq around ARS822 belonging to the class of most affected ARSs. (*) P-value < 0.05; (**) P-value < 0.01.











