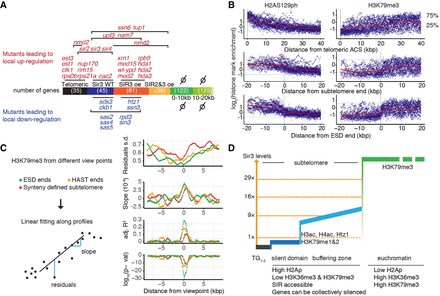
Extension of silent domains reveals new aspects of subtelomeric structures. (A) Localized effects of mutations affecting subtelomeric transcription. The different subtelomeric subdomains are defined according to Sir3 binding. The number of genes within each domain is in parentheses. Mutant names are positioned according to the domain(s) within which the proportion of genes up- or down-regulated (log(FC)>1 or <−1) is significantly elevated (hypergeometric law, with Bonferroni correction n = 703). (B) Distribution of H3K79me3 and H2AS129ph (obtained from Weiner et al. 2015) relative to different subtelomeric viewpoints. Blue lines indicate genome-wide lower and higher quartiles for each mark. The red line corresponds to the LOESS smoothing of histone modification data. (C) Quantification of H3K79me3 transition in function of different genomic viewpoints. Shown are the results of a linear model fitting of the histone mark enrichment data (residuals standard deviation, slopes, R2, and P-values) over 2.35-kb windows every 50 bp. (D) Model depicting how extending silent domains enables to uncover consistent subtelomeric domains delimited by chromatin mark transitions.











