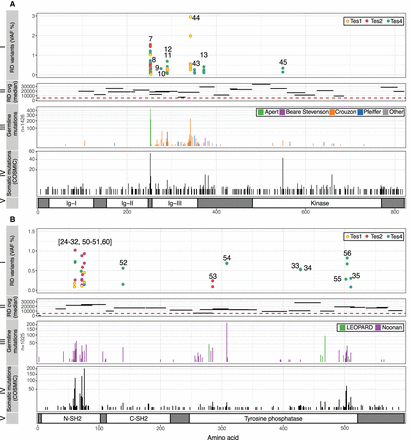
Spontaneous mutations in FGFR2 (A) and PTPN11 (also known as SHP2; B) identified in testicular biopsies. (A, I) Ten validated variants positioned along the amino acid sequence of FGFR2 (x-axis, see panel V), ranging in VAF from 0.06% to 2.95% (y-axis), identified in Tes1D, Tes2F, and Tes4. Numbers correspond to those in Table 1; two different variants (c.870G > C or T) predicted to cause the same p.Trp290Cys substitution (nos. 11, 12) were identified. (II) Relative location and length of amplicons used to sequence main hotspots of FGFR2 are plotted on the x-axis. Median coverage per amplicon is plotted on the y-axis. All amplicons had median coverage above the cut-off (red dashed line) of 5000×. (III) Number of reported constitutional variants encoding amino acid substitutions in FGFR2 associated with developmental disorders (sqrt scale) (updated from Wilkie 2005). (IV) Number of reported somatic amino acid substitutions in FGFR2 in cancer (COSMIC v82). (V) Protein domains of FGFR2. Annotations and protein structure are based on transcript ID NM_000141 and Uniprot ID P21802 (v2017_01), respectively. (B, I) Twenty validated variants positioned along the amino acid sequence of SHP2 (x-axis, see panel V), ranging in VAF from 0.09% to 1.02% (y-axis), identified in Tes1D, Tes2F, and Tes4. (II) Location and size of amplicons used to sequence main hotspots of PTPN11 are plotted on the x-axis. Median coverage per amplicon is plotted on the y-axis. All amplicons except one had median coverage above the cut-off of 5000×. (III) Number of reported constitutional variants encoding amino acid substitutions in SHP2 associated with developmental disorders (sqrt scale). (IV) Number of reported somatic amino acid substitutions in SHP2 in cancer (COSMIC v82). (V) Protein domains of SHP2. Annotations and protein structure are based on transcript ID NM_002834 and Uniprot ID Q06124 (v2017_01), respectively.











