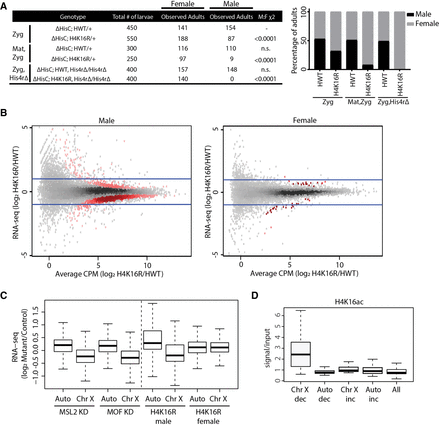
H4K16 promotes hyperexpression of the Drosophila male X Chromosome. (A) Table showing the number of adult females and males resulting from homogeneous vial cultures of 50 first instar larvae of the indicated genotypes (see Supplemental Materials for crosses and complete genotypes). Either only the zygotic (rows 1 and 2) or the maternal and zygotic histones (rows 3 and 4) were of the replication-dependent HWT and H4K16R genotypes. In rows 5 and 6, homozygous deletion of the His4r gene was combined with zygotic, replication-dependent HWT and H4K16R genotypes. The χ2 comparisons were performed using the zygotic HWT male to female results (row 1) as the expected classes. The right panel shows the percentage of viable male (black) and female (gray) adults for H4K16R and HWT. (B) Heatscatter plot of the H4K16R/HWT ratio of RNA-seq signal of individual genes from third instar wing imaginal discs. Statistically different transcripts between H4K16R and HWT males (left panel) or females (right panel) are indicated in red (P < 0.05, edgeR). Blue lines indicate a twofold change. (C) Box plot of RNA-seq signal from autosomes and Chr X after MSL2 or MOF knockdown in male S2 cells (Zhang et al. 2010) and in H4K16R/HWT male and female wing discs on autosomes (Auto) and Chr X. (D) Average enrichment of modENCODE H4K16ac signal from male third instar larvae at 10-kb windows of significantly (P < 0.05) decreased (dec) or increased (inc) transcript expression between H4K16R and HWT males on Chr X and autosomes (Auto) or at all 10-kb windows (GSE49497) (Celniker et al. 2009).











