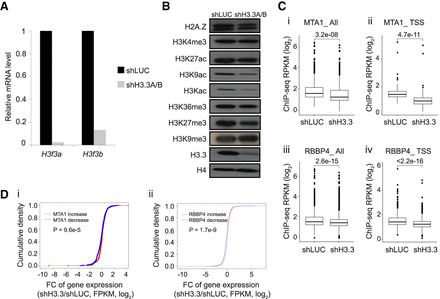
Knockdown of H3.3 leads to changes in global histone marks and a reduction in RBBP4 and MTA1 recruitment. (A) qRT-PCR assays allowing for comparison of H3f3a and H3f3b transcript levels between wild-type (LUC) and H3.3 knockdown cells. (B) Western blot showing changes in levels of histone modifications following knockdown of H3.3. The H3Kac antibody recognizes H3K9ac, H3K14ac, H3K18ac, H3K23ac, and H3K27ac. (C) Box plots showing changes in enrichment for MTA1 genome-wide (i) and ±1 kbp of TSSs (ii), as well as changes in enrichment for RBBP4 genome-wide (iii) and ±1 kbp of TSSs (iv) following knockdown of H3.3. (D) Empirical cumulative distribution for the fold change of gene expression (shH3.3/shLUC), in FPKM (log2), at sites that showed a decrease (blue line) or increase (red line) in MTA1 (i) and RBBP4 (ii) ChIP-seq levels. Genes with MTA1 (i) and RBBP4 (ii) enrichment ±1 kbp from the TSS were included in this analysis. A decrease and increase in ChIP-seq levels after knockdown of H3.3 were defined as log2FC < −0.5 and log2FC > 0.5, respectively.











