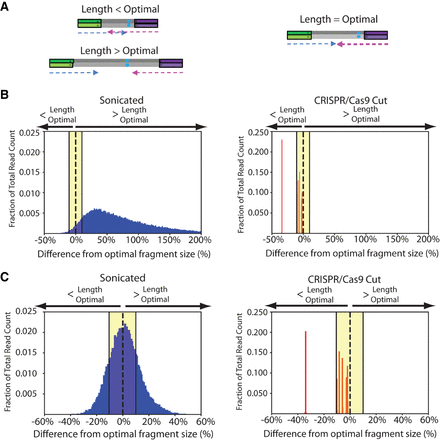
CRISPR/Cas9 fragmentation produces optimal fragment lengths. (A) Sonication produces fragments that are either too short or too long, corresponding to redundant or lost information, respectively. CRISPR-DS produces optimally sized fragments that are perfectly covered by the sequencing reads. (B,C) Comparison of histograms of the insert sizes of two samples prepared with standard-DS (blue, left panels), which uses sonication for fragmentation, and CRISPR-DS (red, right panels), which uses CRISPR/Cas9 digestion for fragmentation. The x-axes represent the percent difference from the optimally sized fragment, e.g., fragment size that matches the sequencing read length after adjustments for molecular barcodes and clipping. Yellow shading highlights the range of fragment sizes that are within 10% difference from optimal size.











