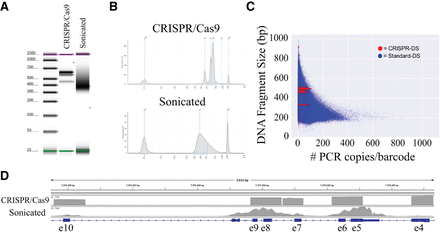
Visualization of sequencing libraries and data prepared with CRISPR-DS and standard-DS. (A) TapeStation gels show distinct bands for CRISPR-DS as opposed to a smear for standard-DS. The size of bands corresponds to the CRISPR/Cas9-cut fragments with adapters. (B) CRISPR-DS electropherograms allow visualization and quantification of peaks for quality control of the library prior to sequencing. Standard-DS electropherograms show a diffuse peak that harbors no information about the specificity of the library. (C) Dots represent original barcoded DNA molecules. Each DNA molecule has multiple copies generated at PCR (x-axis). In CRISPR-DS, all DNA molecules (red dots) have preset sizes (y-axis) and generate a similar number of PCR copies. In standard-DS, sonication shears DNA into variable fragment lengths (blue dots). Smaller fragments amplify better and generate an excess of copies that waste sequencing resources. (D) Integrative Genomics Viewer of TP53 coverage with DCS reads generated by CRISPR-DS and standard-DS. CRISPR-DS shows distinct boundaries that correspond to the CRISPR/Cas9 cutting points and an even distribution of depth across positions, both within a fragment and between fragments. Standard-DS shows the typical “peak” pattern generated by random shearing of fragments and hybridization capture, which leads to variable coverage.











