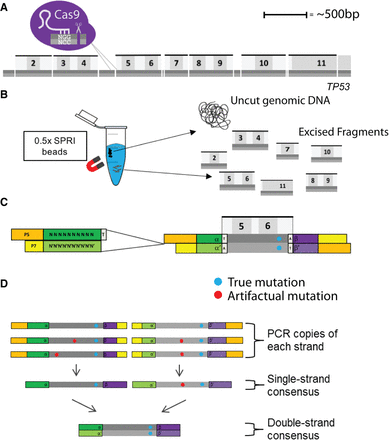
Schematic representation of key aspects of CRISPR-DS. (A) CRISPR/Cas9 digestion of TP53. Seven fragments containing all TP53 coding exons were excised via targeted cutting using gRNAs. Dark gray represents reference strand, and light gray represents the anti-reference strand. (B) Size selection using 0.5× SPRI beads. Uncut, genomic DNA binds to the beads and allows the recovery of the homogenously sized excised fragments in solution. (C) Double-stranded DNA molecule fragmented and ligated with double-stranded DS adapters. Adapters contain 10 bp of random, complementary nucleotides and a 3′-dT overhang. (D) Error correction by DS. After creating single-strand consensus sequence (SSCS) reads, SSCS reads derived from the same original DNA molecule are compared with one another to create a double-strand consensus sequence (DCS). Only mutations found in both SSCS reads are counted as true mutations in DCS reads.











