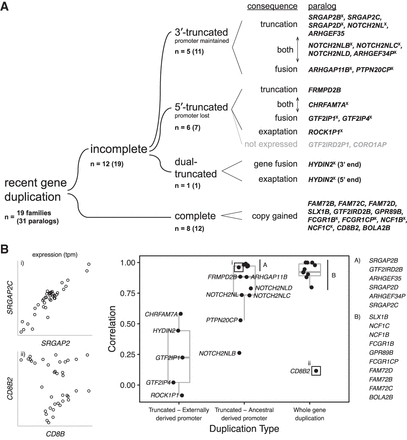
Transcriptional fates of human-specific duplicate genes and expression correlation between ancestral and duplicate gene copies. (A) We classify 19 gene families (31 duplicate paralogs in GRCh38) by the transcriptional characteristics of the duplicate genes. In eight of 19 gene families and 12 of 31 paralogs, the duplication includes the complete gene (with respect to the canonical isoform). More common are incomplete gene duplications, of which five of 12 gene families and 11 of 19 paralogs are 3′ truncated (whereby the ancestral promoter is maintained in the duplicate gene), while six of 12 gene families and seven of 19 paralogs are 5′ truncated (whereby the ancestral promoter is lost). The outcomes of such truncated duplications can be simply shortened versions of the ancestral gene (“truncation”) or transcript fusion with adjacent sequence (“fusion”), and often both are observed. For 5′ truncations, we also observe the phenomenon of exaptation of upstream exons and regulatory elements, which provide a new promoter for what would presumably be otherwise transcriptionally silent genes (two of 19 gene families). Note that duplicates of GTF2IRD2 are classified as both complete and incomplete. (B) We estimated expression similarity between ancestral and duplicate copies by calculating the pairwise correlation of the median expression levels across GTEx tissues. Duplicate genes whose promoters were included in the human-specific SD show expression patterns that are more similar to their ancestors than those that acquire it from new sequence.











