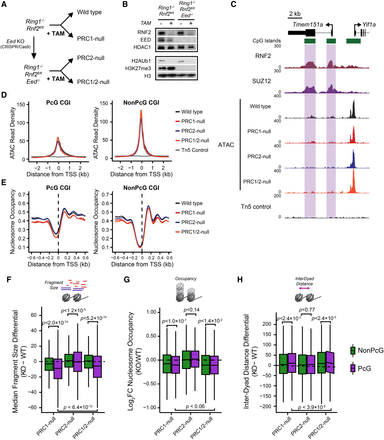
PRC1 and PRC2 do not function redundantly to shape the chromatin landscape at PcG-occupied gene promoters. (A) A schematic detailing the strategy to ablate PRC1 and/or PRC2 in mouse ESCs. (B) A Western blot analysis for RNF2, EED, H3K27me3, and H2AK119ub1 for Ring1−/−;Rnf2fl/fl and Ring1−/−;Rnf2fl/fl;Eed−/− ESCs with or without tamoxifen (TAM) treatment after 72 h. (C) A genome screenshot for Ring1−/−;Rnf2fl/fl and Ring1−/−;Rnf2fl/fl;Eed−/− ATAC-seq signal before and after TAM at PcG-occupied CGIs (highlighted in purple) and a non-PcG CGI promoter. (D) A metaplot analysis for Ring1−/−;Rnf2fl/fl and Ring1−/−;Rnf2fl/fl;Eed−/− ATAC-seq before and after TAM treatment at PcG-occupied (n = 4020) or non-PcG CGI promoters (n = 10,251), centered on TSSs. (E) A metaplot analysis for Ring1−/−;Rnf2fl/fl and Ring1−/−;Rnf2fl/fl;Eed−/− NucleoATAC-derived nucleosome occupancy score before and after TAM treatment at PcG-occupied or non-PcG CGI promoters, centered on TSSs. (F) A box plot comparing the change in median Tn5-tagmented fragment sizes for PcG and non-PcG CGI promoters between Ring1−/−;Rnf2fl/fl and Ring1−/−;Rnf2fl/fl;Eed−/−ATAC-seq data sets before and after TAM treatment. (G) A box plot quantifying the log2 fold change (log2FC) in NucleoATAC-derived nucleosome occupancy for PcG and non-PcG CGI promoters in Ring1−/−;Rnf2fl/fl and Ring1−/−;Rnf2fl/fl;Eed−/− ATAC-seq data sets before and after TAM treatment. (H) A box plot comparing the difference in median inter-dyad distances for PcG and non-PcG CGI promoters in Ring1−/−;Rnf2fl/fl and Ring1−/−;Rnf2fl/fl;Eed−/− ATAC-seq data sets before and after TAM treatment.











