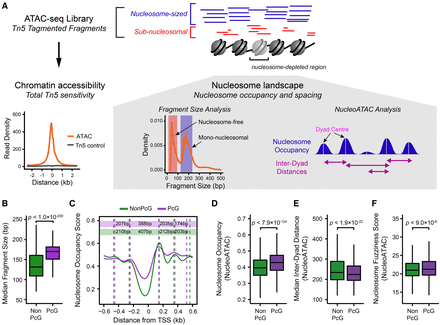
Characterization of the nucleosome landscape at Polycomb-occupied promoters. (A) A schematic detailing the approach to analyze nucleosome landscape features from ATAC-seq data. The cleavage of Tn5 hypersensitive DNA (accessible DNA) by Tn5 generates DNA fragments that broadly reflect either mononucleosomal fragments (blue) or nucleosome-free fragments (red). The total count of fragments represents total chromatin accessibility at a given loci, and the fragment size distribution allows the examination of qualitative features of Tn5 sensitivity, such as nucleosome occupancy or positioning using either the median fragment size for a gene promoter or the quantification of nucleosome occupancy signal using the software package NucleoATAC (Schep et al. 2015). After identifying nucleosome positions using NucleoATAC, individual nucleosome dyad centers can then be identified, and the distance between neighboring dyad centers can be calculated. (B) A box plot comparing the median ATAC-seq fragment sizes for PcG-occupied (n = 4020) or non-PcG (n = 10,251) CGI promoters. PcG-occupied promoters tend to have larger fragment sizes consistent with an enrichment for nucleosomal-sized fragments. (C) A metaplot for PcG-occupied or PcG-free promoters depicting nucleosome occupancy signal extracted from ATAC-seq data using NucleoATAC, centered on TSSs. The average dyad center for each nucleosome position is marked by dashed lines, and the distance between each nucleosome position is included in the colored rectangles: (purple) PcG; (green) NonPcG. (D) A box plot comparing the NucleoATAC-derived nucleosome occupancy score within PcG-occupied or PcG-free promoters. (E) A box plot comparing the median inter-dyad distances within PcG-occupied or PcG-free promoters. Distances were calculated between the centers of neighboring dyad positions identified by NucleoATAC. (F) A box plot comparing the median fuzziness score for nucleosomes identified by NucleoATAC within PcG-occupied or PcG-free promoters.











