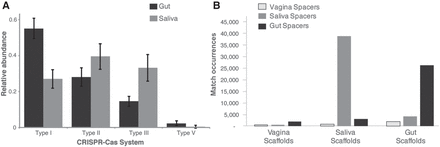
Diversity and distribution of CRISPR-Cas systems and CRISPR spacer targets (mobile elements). (A) Distribution of CRISPR-Cas system types in the gut (gray) and saliva (black) samples in this study (I, II, III, V). Vaginal samples are not shown due to low bacterial diversity. The black lines indicate standard deviation. (B) Frequency of matches between spacers and scaffolds. This graph shows the total number of matches by body site (outlined gray, vaginal spacers; light gray, saliva spacers; and dark gray, gut spacers). There were a total of 78,054 matches between spacers and all pregnancy scaffolds. Overall, we detected 3254 spacer types from vaginal samples, 36,477 spacer types from gut samples, and 36,279 spacer types from saliva samples.











