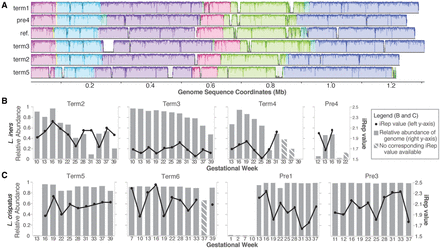
Comparative genomics of Lactobacillus spp. and estimated replication rates. (A) Multiple genome alignment of five L. iners genomes recovered through metagenomics along with the reference strain DSM 13335 (see Methods). A modified alignment created in Mauve shows the shared genomic context, as well as genomic islands unique to a strain (white areas within a conserved block). (B,C) Relative abundance and estimated genome replication values (iRep) for L. iners and L. crispatus, respectively.











