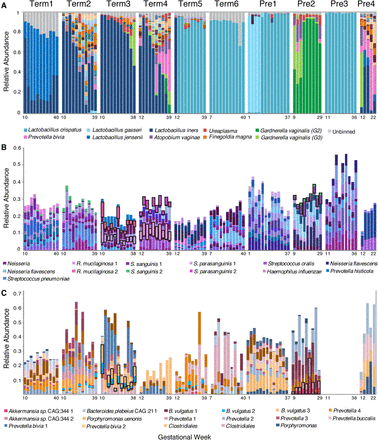
Community structure of the pregnancy microbiome over time in 10 subjects. (A) Relative abundance of vaginal genome bins (y-axis). Abundance was estimated from the number of reads that mapped to each bin and normalized by the length of the bin. The top 10 most abundant vaginal taxa are displayed (see key at bottom). (Unbinned) Sequences that were not assigned to a classified genome bin. (B,C). Relative abundance (y-axis) of the top 50 most abundant taxa across all subjects in saliva (B) and gut (C) samples, respectively. Gut samples from subject Pre3 were not available. Species abundance was estimated from the average read counts of single-copy ribosomal protein (RP) sets (at least one of 16), summed over scaffolds sharing RPs clustered at 99% amino acid identity. Each taxon is represented by a distinct color (see key for selected taxa at bottom) and is classified at the most resolved level possible. Co-occurring strains of Rothia mucilaginosa, Streptococcus sanguinis, and Streptococcus parasanguinis in B, and of Bacteroides vulgatus and Akkermansia spp. CAG:344 in C are highlighted with black boxes.











