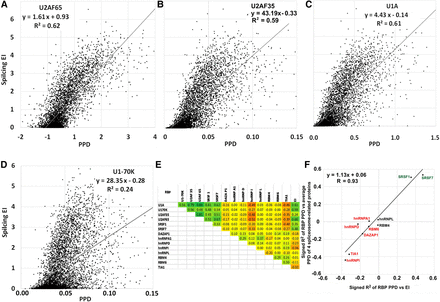
Correlation between splicing (EI) and RBP-exon binding in vitro. A library of the 5560 exon sequences plus short flanks was incubated with a HeLa nuclear extract and immunoprecipitated to pull down bound RNA molecules, which were quantified by deep-sequencing. PPD is the proportion of the library bound for each sequence, including correction for input proportion and the amount of RNA recovered. (A) U2AF65 (U2AF2); (B) U2AF35 (U2AF1); (C) U1A; (D) U1-70K. A and B are required for U2 snRNP binding, and C and D are components of the U1 snRNP. Both snRNPs are part of the initial spliceosome. (E) Correlation in binding among RBPs. Numbers shown are signed R2 values. Positive, negative, and no correlations were found. Note that the four spliceosome-related proteins (U1A, U1-70K, U2AF65, U2AF35) positively correlated with SRSF1 and SRSF7 and negatively correlated with DAZAP1, hnRNP I (PTB), and TIA1 binding (see Supplemental Fig. S11 for additional scatter plots). Correlations with splicing (EI, last column) are also shown. (F) Individual RBPs may promote or prevent the formation of a functional exon definition complex. On the x-axis are plotted the correlations (as signed R2 values) between the binding (PPDs) of individual RBPs to the mutant exons and splicing. On the y-axis are plotted the correlations between the binding (PPDs) of individual RBPs to the mutant exons and the average binding (PPDs) of the four spliceosome assembly proteins U2AF35, U2AF65, U1A, and U1-70K; these values were taken as an indicator of exon definition complex formation. This correlation of correlations plot allows a visualization of the positive relationship between the promotion (repression) of exon definition by an RBP and the promotion (repression) of splicing by that RBP. The correlations do not show causality but are consistent with that idea.











