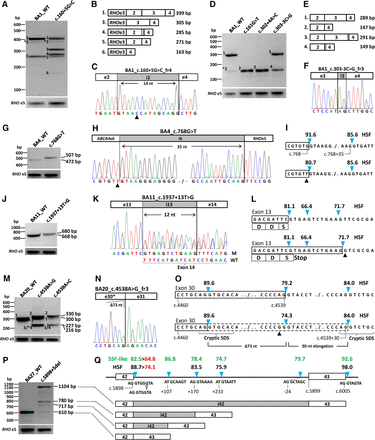
Overview of splice defects due to nine noncanonical splice site variants in ABCA4. All wild-type and mutant midigenes were transfected in HEK293T cells and their RNA subjected to RT-PCR. (A,B) RT-PCR for wild-type and c.160+5G>C mutant BA1 midigene showed complex splicing defects when using primers in RHO exon 3 and ABCA4 exon 4. Five defects (fragments 2 through 6) were observed next to the wild-type fragment 1. Asterisks denote fragments for which we obtained no sequence information. The RT-PCR products showed skipping of exon 2, exon 3, or exons 2 and 3. (C) Sequencing of pGEM-T-cloned fragment 4 revealed a 14-nt insertion due to the activation of a cryptic splice donor site in intron 2 and the absence of exon 3. The triangle points to the +5G>C variant. (D,E) RT-PCR products of wild-type and mutant BA1 containing NCSS variants residing at splice sites of exons 3 and 4. Primers in ABCA4 exons 2 and 4 reveal exon 3 skipping and/or exon 4 elongation. Note that the wild-type vector BA1 shows exon 3 skipping (fragment 5), which is more pronounced when using a forward RHO exon 3 primer (A) compared to using a forward ABCA4 exon 2 primer (D). (F) Sanger sequence of gel-purified fragment 3 shows a 2-nt elongation at the 3′ end of exon 4 due to the activation of a cryptic splice acceptor site in intron 3. Sanger sequencing results for all fragments without an asterisk are provided in Supplemental Figure S3. (G,H) RT-PCR and Sanger sequencing of RNA extracted from c.768G>T mutant BA4 midigene showed a 35-nt exon 6 elongation. (I) Human Splice Finder (HSF) splice site scores (blue arrowheads) for wild-type and mutant sites. (J,K) RT-PCR, gel analysis, and Sanger sequencing showed the wild-type product and a 12-nt exon 13 elongation due to c.1937+13T>G. M and WT denote mutant and wild-type splice products. (L) Details of the exon elongation defect. As represented by the HSF prediction software, the scores of the normal and intronic splice donor sites (SDSs) are not changed due to this variant. Most of the RT-PCR product consists of the exon elongation which introduces a premature stop-codon. (M–O) Variants at the penultimate position of exon 30 (c.4538A>G and c.4538A>C) resulted in a correct mRNA (fragment 1), a 30-nt exon 30 elongation due to the presence of a strong cryptic intronic splice donor site (fragment 2), a deletion of 73 nt of the 3′ end of exon 30 (fragment 3), and a 30-nt exon 30 elongation in combination with a 114-nt deletion of the 5′ end of exon 30 (fragment 4). Fragments 3 (227 bp) and 4 (216 bp) could not be resolved. The exon 30 deletions are the results of cryptic splice donor and acceptor sites that overlap at exonic positions c.4465–4468 (Supplemental Fig. S2, BA20). (P) RT-PCR of BA27 carrying the c.5898+5del variant resulted in at least two exon 42 elongation products of 717 and 780 bp, as well as intron 42 retention. (Q) The exon elongation products were due to the reduction of the strength of the exon 42 splice donor site, as shown with red numbers, in combination with the presence of nearby cryptic intronic splice donor sites with strong predicted scores. Only HSF and SpliceSiteFinder (SSF)-like scores are provided. Note that SSF-like is the only software that predicts the rare splice donor sites (positions c.5898+108 and c.5899−23) that contain the canonical GC sequence instead of GU. Intron 42 retention likely is due to the very strong splice donor site (HSF 98.0) flanking exon 43. The open arrowhead denotes the 1-bp deletion.











