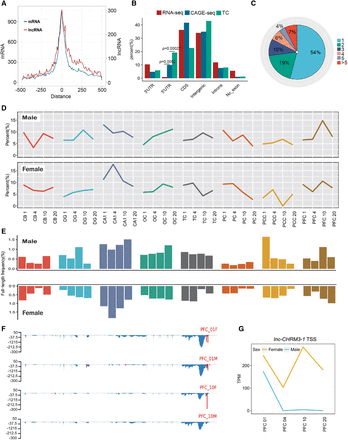
CAGE-seq characterization of alternative promoter usage and full-length frequency of lncRNAs. (A) TC number distribution around the annotated TSS of known mRNAs (turquoise) and lncRNAs (red) identified in this study. (B) Genomic region distribution of RNA-seq reads (control), CAGE-seq reads, and TCs to view the CAGE signal enrichment. Enrichment P-values were labeled for the 5′ UTR (Fisher's exact test). (C) Pie chart of percentage for lncRNA genes with one and more promoters. (>5) lncRNA genes with six and more promoters. (D) Dynamics of alternative promoter profiles during macaque brain development and aging. Alternative TC reads divided by total TC reads for each of the 64 samples were calculated and plotted. Male and female samples were separately plotted. (E) Dynamics of full-length frequency profiles of all lncRNAs in all 64 brain samples. Full-length frequency is indicated by the detected fraction of lncRNAs with polyadenylation and 5′-capping. The x-axis label is the same as in D. (F) Distribution of RNA-seq reads density (blue) and CAGE-seq TCs (red) along the lnc-CHRM3-1 lncRNA. y-axis indicates the normalized density for RNA-seq and CAGE-seq. (G) Line plot of lnc-CHRM3-1 TSS density in PFC samples. The TSS density was represented by TPM.











