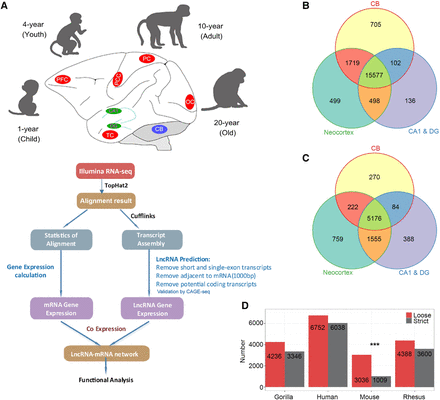
A comprehensive catalog of lncRNA genes in rhesus monkey brain. (A) Illustration of the experimental design and bioinformatics analysis pipeline for the identification and functional annotation of lncRNA genes expressed in macaque brain. Macaque brain regions used in this study were colored in red (neocortex), green (hippocampus), and blue (cerebellum). See Methods for more details. (B,C) Venn diagram of detected mRNA (B) and lncRNA (C) genes in neocortex, hippocampus (CA1 & DG), and cerebellum. (D) Number of NONCODE lncRNAs in gorilla, human, mouse, and rhesus that are homologous to macaque brain lncRNAs identified in this study with a loose (E-value < 1 × 10−3) and strict (E-value < 1 × 10−10) threshold by BLASTN. Alignment to mouse showed a significant decrease with the strict threshold. (***) P-value < 0.0001 by Fisher's exact test, human as background.











