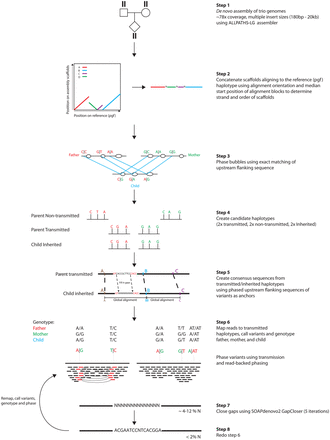
Assembly of 100 full MHC haplotypes. Schematic showing the construction of MHC haplotypes. Genomes in trios are de novo assembled using ALLPATHS-LG (step 1). Scaffolds larger than 50 kb mapping to the MHC are extracted and concatenated, creating diploid consensus scaffolds (step 2). Bubbles in the alignment graphs for individuals in the trio are mapped uniquely within the trio by exact matching of the sequence upstream of the bubbles (step 3). Global alignment between phased bubbles is used to create a consensus sequence between transmitted parental and inherited child haplotype sequences (steps 4 and 5). Reads from parents and child are then mapped to the consensus sequence, genotyped, and phased (step 6), gaps are closed (step 7), and reads are mapped again for another iteration of mapping, genotyping, and phasing (step 8).











