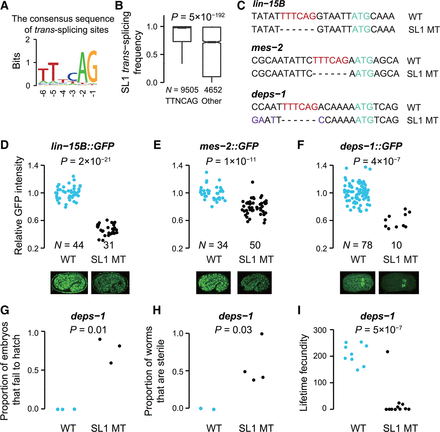
Knocking out the consensus sequence of trans-splicing sites leads to reduced translational efficiency. (A) The consensus sequence of SL1 trans-splicing sites in C. elegans (TTNCAG). (B) The consensus sequence significantly increases the efficiency of SL1 trans-splicing (P = 5 × 10−192, Mann-Whitney U test). (C) The sequences of wild type (WT) and SL1 trans-splicing mutants (SL1 MT) around the trans-splicing sites are shown for lin-15B, mes-2, and deps-1. The consensus sequences of trans-splicing sites are marked in red and the start codons are marked in cyan. Silent mutations were made (in purple) to prevent Cas9 from recutting. (D–F) The protein abundances of LIN-15B (D), MES-2 (E), and DEPS-1 (F) were quantified by the fluorescence intensity of GFP. P-values were given by the Mann-Whitney U test. Because knocking out the trans-splicing site of deps-1 frequently resulted in embryonic lethality and a complete loss of GFP signal, only embryos with GFP signal were used for quantification. An image example of each strain is shown. (G–I) Knocking out the trans-splicing site of deps-1 induced partially penetrant embryonic lethality (G), reduced fertility (H), and decreased lifetime fecundity (I). Each dot represents the result of an independent experimental replication. P-values were given by the t-test.











