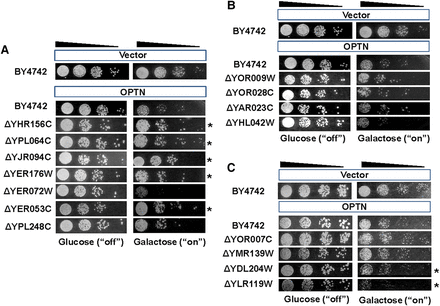
Validation of the genome-wide screening data for OPTN. Barcode counting was used to screen OMIM-yeast genetic interactions. From the Bar-seq data analysis and Z-score distributions, three groups of yeast genes were chosen for spotting assays: group 1, toxicity suppressors; group 2, no effects; and group 3, toxicity enhancers. OPTN toxicity was analyzed with spotting assays in the wild-type yeast strain (BY4742), toxicity-suppressing gene deletion strains (A), no-effect gene deletion strains (B), and toxicity-enhancing gene deletion strains (C). pAG425GAL-ccdB was used as the empty vector control. Twenty-four yeast deletions were tested. Representative results are shown. Asterisks indicate yeast deletions that suppressed or enhanced OPTN toxicity.











