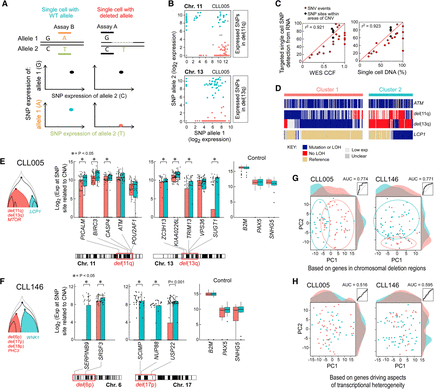
Integrated analysis of DNA- and RNA-level information by simultaneous detection of mutation, deletion, and gene expression in single cells. (A) Schema of deletion call based on expression of heterozygous SNPs. Dashed line indicates deleted region. A deletion was called when a cell expresses only one allele in the deleted region while expressing similar levels of both alleles for SNPs outside the deleted region. (B) Examples for deletion calls in CLL005 single cells for subclonal deletions in Chromosomes 11 and 13. Each point represents a cell. Orange indicates cells with no detectable expression from the deleted allele are inferred to harbor the deletion; blue, cells with expression from both alleles or the deleted allele are inferred to lack the deletion. (C) Mutation and chromosomal deletion frequency detected from single-cell RNA from five CLL samples show high correlation with CCF (r2 = 0.921) and with mutation frequency per single-cell DNA analysis (r2 = 0.923). (D) Mutation and chromosomal deletion detection in single-cell RNA from sample CLL005 enables reconstruction of phylogeny. (E,F) Genes within the chromosomal deletion regions exhibit significantly lower expression based on a one-sided Wilcoxon rank-sum test in cells inferred to harbor the deletions (cluster 1; orange) compared with cells not harboring the deletions (cluster 2; blue). Housekeeping genes are not significantly differentially expressed among single cells from the two clusters. (G) PCA of single cells from CLL005 or CLL146 based on gene expression for genes within chromosomal deletion regions leads to separation of cells by genetic subpopulation. Linear discriminant analysis achieves elevated ROC AUCs of 0.774 for CLL005 and 0.771 for CLL146. (H) PCA of single cells from CLL005 or CLL146 based on gene expression for genes driving aspects of transcriptional heterogeneity (23 genes for CLL005, 33 for CLL146) fails to separate cells by genetic subpopulation. Linear discriminant analysis does not perform substantially better than random, achieving ROC AUC of 0.516 for CLL005 and 0.595 for CLL146.











