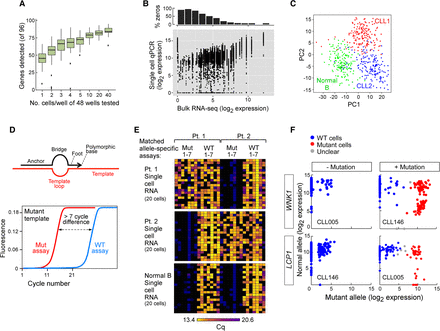
Establishment of a targeted RNA-based approach to perform integrated, targeted, and multiplexed detection of somatic mutations and gene expression in single cells. (A) Number of genes detected from 96 genes using cell numbers ranging from one to 40 cells per well by the targeted approach illustrated in Figure 1A, right panel. (B) Correlation between gene expression derived from a bulk RNA-seq and single-cell targeted approach in CLL005 (analyses of other individual CLL samples and in aggregates in Supplemental Fig. S7). (C) Gene expression of a set of 96 genes in single cells distinguishes normal (CD19+) and CLL-B cells by principal component analysis (PCA). Single cells were derived from one normal healthy donor and two CLL patients. (D, top) SuperSelective primer design (Vargas et al. 2016) was used for mutation detection. The primer contains a long 5′-anchor sequence that binds strongly to template strands, a short 3′-foot sequence that includes an interrogating nucleotide complementary to the corresponding nucleotide in a mutant template (but mismatches the corresponding nucleotide in a wild-type template), and a linking bridge sequence. (Bottom) Schema of a successful targeted mutation detection assay, in which distinct paired assays (wild-type and mutant allele) were designed for detection of a single mutation. (E) Heat map of seven patient-specific mutation detection assays (with seven matched wild-type assays) performed on cDNA derived from 20 single cells from normal CD19+ B cells or two CLL-B cells, with detection measured as the number of cycles to achieve the detection threshold (Cq). (F) Examples for mutation calls in single cells with or without mutations LCP1 and WNK1. Cells were called wild-type (WT; blue), mutant (red), or unclear (gray).











