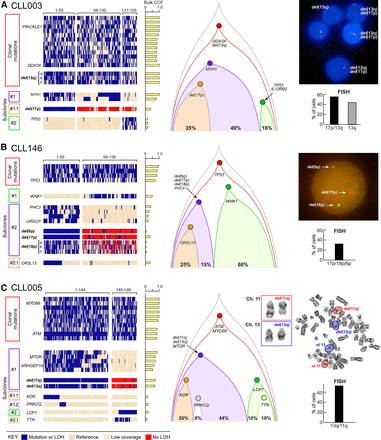
Reconstruction of tumor phylogeny in CLL from single-cell DNA analysis. (A–C) Detection and clustering of somatic mutations and chromosomal deletions (rows) for single viable CD19+CD5+ cells (columns) from CLL003 (A), CLL146 (B), CLL005 (C; for analysis of CLL096 and CLL032, see Supplemental Fig. S4A,B). (Left) Blue indicates the presence of mutations (sSNVs) or chromosomal deletion; beige, the absence of sSNVs; red, the absence of chromosomal deletion. (Middle) Clonal architecture for CLL003, CLL146, and CLL005 derived from single-cell DNA analysis. (Right) FISH hybridization (A,B) and karyotyping images (C) as validation of the single-cell chromosomal deletion analysis with percentage of positive FISH cells enumerated from 100 cells. Sensitivity of probes used in the study was confirmed by hybridization with PBMC from a normal donor (Supplemental Fig. S4C).











